Reconstitution of a minimal mechanotransduction system
Vincent L.M.F. Louis
v.l.m.f.louis@tudelft.nl
Bio
Hi, my name is Vincent, I'm from Paris. I have a background in bioengineering and chemistry. After gaining some research experience in Paris and abroad, I joined the artificial cell adventure at TU Delft. I am particularly interested in reconstructing a mechanotransduction system at the single-molecule level, for an artificial cell. I like cinema, and history.
Project summary
Cells actively sense and respond to mechanical forces. Integrins and their partners, such as talin, and the actin cytoskeleton form the core of this force-sensing machinery. We aim to rebuild this minimal system using optical tweezers and TIRF microscopy, to ultimately give artificial cells mechanosensing abilities. I like cinema and history.
Introduction
Living cells are often depicted as simple spherical containers floating in their environment. In reality, in vivo, they rarely resemble this ideal geometry: cells are constantly compressed, stretched, and sheared as they grow, divide, and mature. They are far more than just passive containers: they are mechanical objects capable of sensing the forces acting upon them, both external and internal, and responding by reshaping themselves.
At the heart of this mechanical dialogue, at the interface between the extracellular matrix and the cytoplasm, lie the integrins: transmembrane proteins that span the lipid membrane and mechanically link extracellular ligands to the cytoskeleton.
This connection, however, is not direct. Just beneath the inner membrane lies a dense layer of adaptor proteins that mediate the coupling. Among them is talin, a mechanosensitive protein that, when stretched, exposes new binding sites for both partners and the actin cytoskeleton.
The full complexity of this system exceeds the capabilities of a prototype artificial cell, but it suggests a set of minimal design principles:
-
an extracellular matrix,
-
a transmembrane protein anchored to it,
-
a stretchable protein that can bind to the cytoplasmic tail of the transmembrane protein,
-
other adapter proteins, or the cytoskeleton itself, that can be recruited when the adapter is stretched.
Although the components of this interface have been studied in isolation, our aim is to bring them together and recreate stretching under more realistic conditions: within a biomimetic lipid environment, in the presence of the cytoplasmic tail of a simplified integrin, and alongside the cytoskeleton.
To achieve this, we are building a protein-tethered bilayer lipid membrane, essentially a lipid bilayer supported by a polymer cushion, into which we want to incorporate an integrin anchored to the surface. Our next step is to establish a single-molecule approach: dragging a talin hook, attached to a DNA handle, across the surface of the bilayer. The objective is to capture the integrin tail with this talin hook, unfold it by pulling, recruit downstream proteins, and visualize the process in real time with TIRF microscopy.
In the longer term, to build an artificial cell with mechanosensing abilities, this minimal system must also function once encapsulated. This presents several challenges. Whereas integrins are typically purified from blood platelets, here they must instead be expressed in E. coli and purified, or synthesized in situ (e.g., using PURE) alongside their partners: talin and the actin cytoskeleton. Moreover, the integrins should spontaneously insert into the membrane, and the artificial cell should be deformable enough to reproduce stretching.
Some articles:
https://doi.org/10.1016/j.bpj.2016.08.051
https://doi.org/10.1038/s41467-024-49222-z
https://doi.org/10.3390/membranes11070499
https://doi.org/10.1016/j.cell.2024.04.049
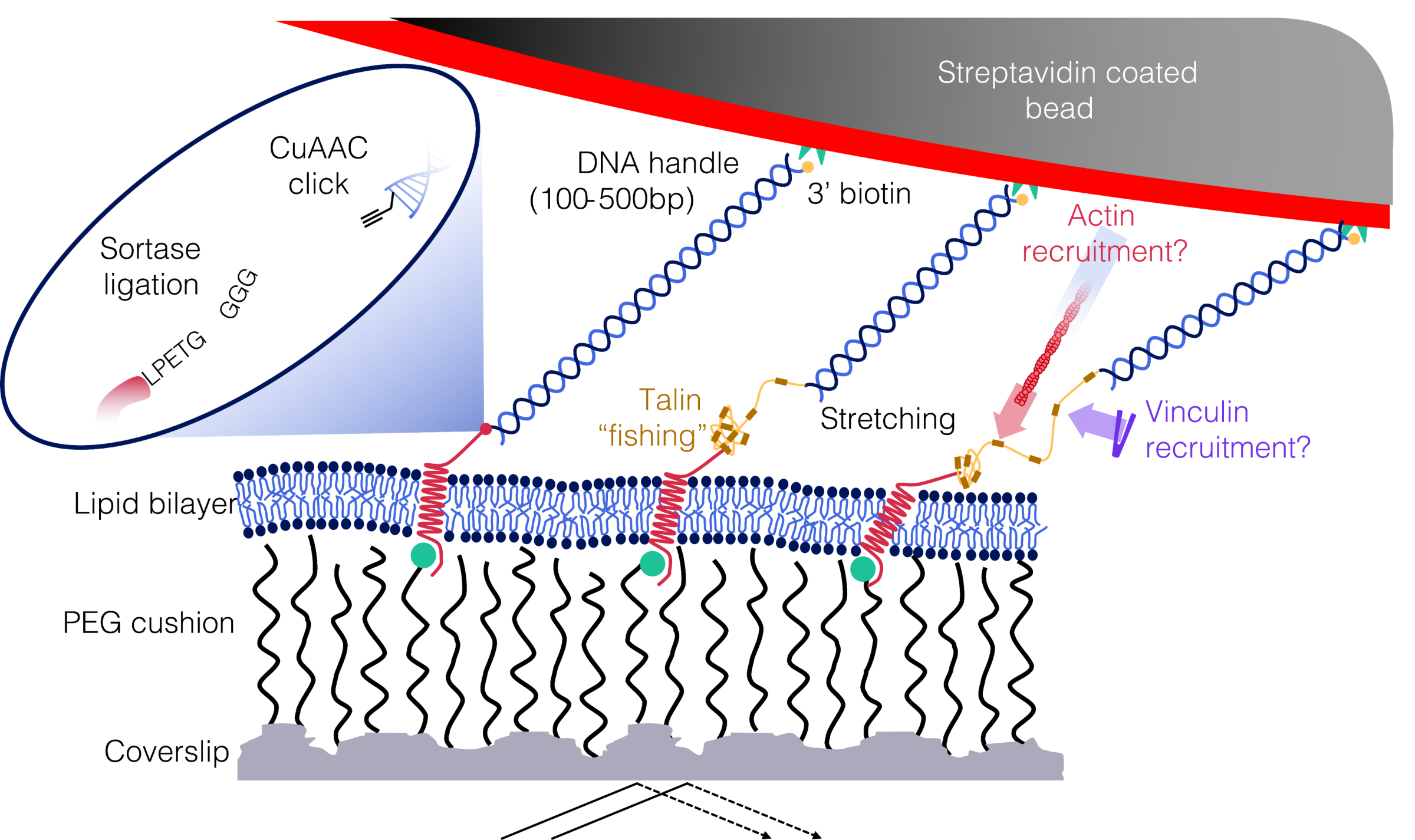
Strategy for capturing and unfolding talin in a biomimetic environment with a single-bead optical tweezer
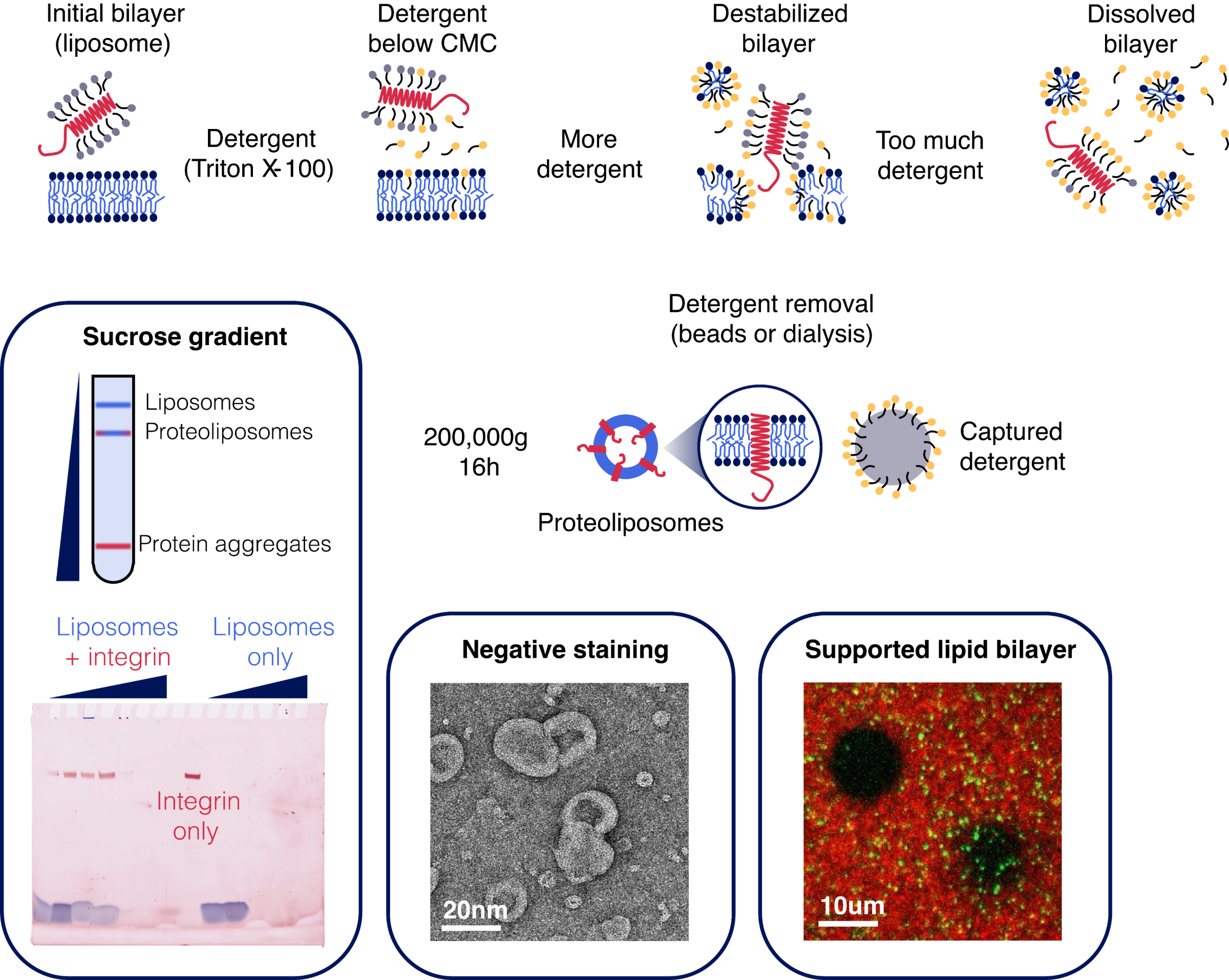
The one-pot detergent removal method: a timeless approach to proteoliposome formation
The aim is to generate a supported lipid bilayer (SLB) containing transmembrane integrin. The most common quality controls for proteoliposome reconstitution include sucrose gradient centrifugation and electron microscopy (in this case, negative staining). The integrity of the resulting SLB is typically evaluated using FRAP. A key limitation lies in determining whether the protein is truly inserted across the membrane, rather than merely adsorbed onto its surface. For channels, this can be addressed with functional assays, for example, by measuring fluorescence fluctuations of a dye trapped within liposomes. In contrast, for structural proteins such as integrins, the challenge is greater and usually requires increasing imaging resolution to directly resolve the bilayer.
Design of a simplified, minimal integrin.
Integrins are too large to express in E. coli, but simplified designs make it possible. The large and complex head is removed and replaced either by a polyhistidine to bind to an extracellular matrix mimicked by an NTA-functionalized surface, or more realistically by an RGD binding domain. Also, we only keep the beta subunit, which binds to the cytoskeleton.
Simplified view of the two states of an integrin heterodimer
The bent/extended state refers to the head on the extracellular matrix side, which is extended and capable of binding to its ligand, or on the opposite bent and unbound. The closed/open state refers to the state of the two transmembrane and cytoplasmic legs. The hindrance created by the closed state hides the residues in the beta integrin tail that bind to the cytoskeleton and its adapter partners.





